MANHATTAN — Kansas State University researchers have led a newly published study that is helping unlock some of the secrets of specific cellular messengers called exosomes. The work could one day lead to therapeutic interventions against devastating neurodegenerative diseases and debilitating spinal cord injuries.
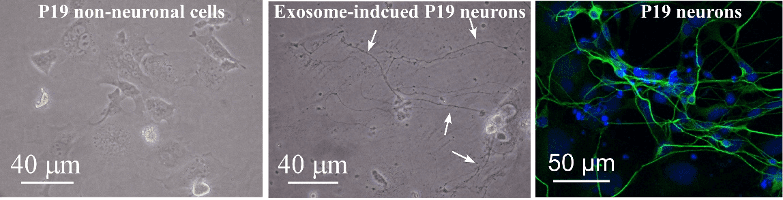
“In our new manuscript, we demonstrated the generation of neurons using exosomes in culture,” said Meena Kumari, associate professor in the College of Veterinary Medicine. “It is our hope that the ability to generate nerve cells in a dish can one day give hope to those who suffer spinal cord injuries and to those millions who are diagnosed with neurodegenerative diseases.”
Kumari led a multidisciplinary team that has published the results of the study in “Exosomes induce neurogenesis of pluripotent P19 cells” in Stem Cell Reviews and Reports.
P19 cells are pluripotent embryonal carcinoma cells, and exosomes are small membrane-bound structures released by cells that contain unique cargo of small RNAs, proteins, lipids and metabolites, Kumari said.
“We can’t see them exosomes with the naked eye, but they are very important in cell-cell communication and can have profound impact on the biology of cells that engulf them,” Kumari said. “Exosomes’ cargo often reveal their parent cell origin’s effects on the recipient cells in many ways.”
In the study, the researchers reported that exosomes released from P19 neurons were able to induce differentiation of P19 non-neuronal cells into neurons, Kumari said. The team used RNA sequencing to identify small RNAs that could drive the differentiation of non-neuronal cells to neurons. Small RNAs encompass several classes of functional noncoding RNAs including microRNAs that are encased in exosomes released from P19 neurons.
MicroRNAs can modulate expression of several genes simultaneously. As a result, noncoding RNAs have the ability to transform the genetic landscape of cells that engulf them. Kumari and the team identified a number of noncoding RNAs that potentially contributed to differentiation of P19 non-neuronal cells to neurons.
Kumari’s lab is conducting further experiments that employ various strategies, including CRISPR/CAS9. The researchers want to decode and understand the precise functional role of P19N exosomal noncoding RNAs in modulating gene expression to drive the differentiation of P19 non-neuronal cells to neurons.
K-State co-authors on the study included Antje Anji, research associate professor of anatomy and physiology; Takashi Ito, professor of chemistry; and David A. Meekins, instructor of biochemistry and molecular biophysics.
Other researchers on the project included Briana Anderson, visiting scholar from Langston University in Oklahoma and senior in biology who spent a summer conducting research in Kumari’s lab; Feroz Akhtar, postdoctoral fellow from Texas A&M University; and Srinivas Mummidi, professor at Texas A&M University-San Antonio.
Funding for this work was provided by the K-State department of anatomy and physiology and through intramural grants in the College of Veterinary Medicine.